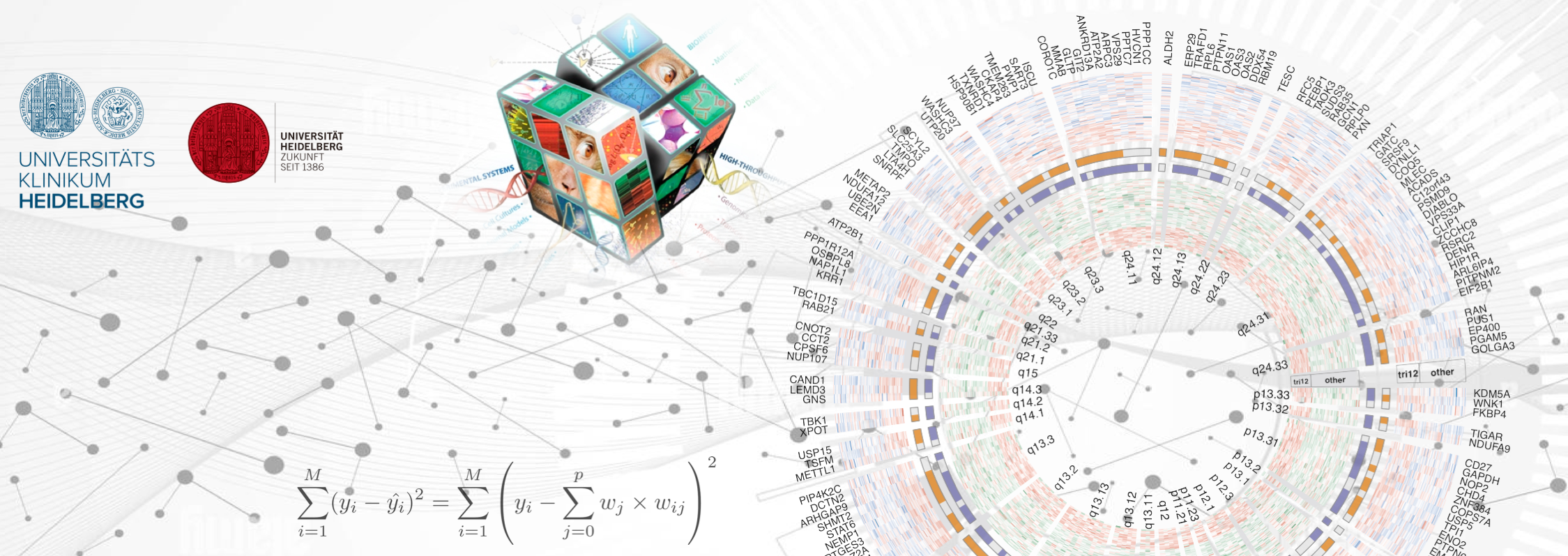
Computational Omics and Precision Oncology
Fighting cancer and solving the mystery of life through the power of multi-omics and data science
Research
We are a computational group embedded in a clinical setting. We focus on developing and applying statistical and machine learning tools for mining complex biological and clinical data, to identify cancer biomarkers and drug targets. We work closely with physicians and clinician scientists to translate our search outputs into better patient care.

SMART-CARE Consortium
We are part of the SMART-CARE consortium, which aims to develop novel systems medicine approaches to battle cancer recurrence, using the integration of high-quality proteome and metabolome mass spectrometry data with other ‘omics and clinical data. The role of our group in the consortium is to provide innovative and robust computational solutions for mining and integrating mass-spectrometry data, in order to discover cancer biomarkers. Read more about SMART-CARE.

MMPU Group
The Molecular Medicine Partnership Unit (MMPU) is a joint venture between the Medical Faculty of the University of Heidelberg and the European Molecular Biology Laboratory (EMBL). We are part of the Systems Medicine of Cancer Drugs MMPU group. Together with the groups of Wolfgang Huber and Sascha Dietrich, we aim to understand intra- and inter-patient heterogeneity of response to anti-cancer drugs, a major clinical and scientific challenge.
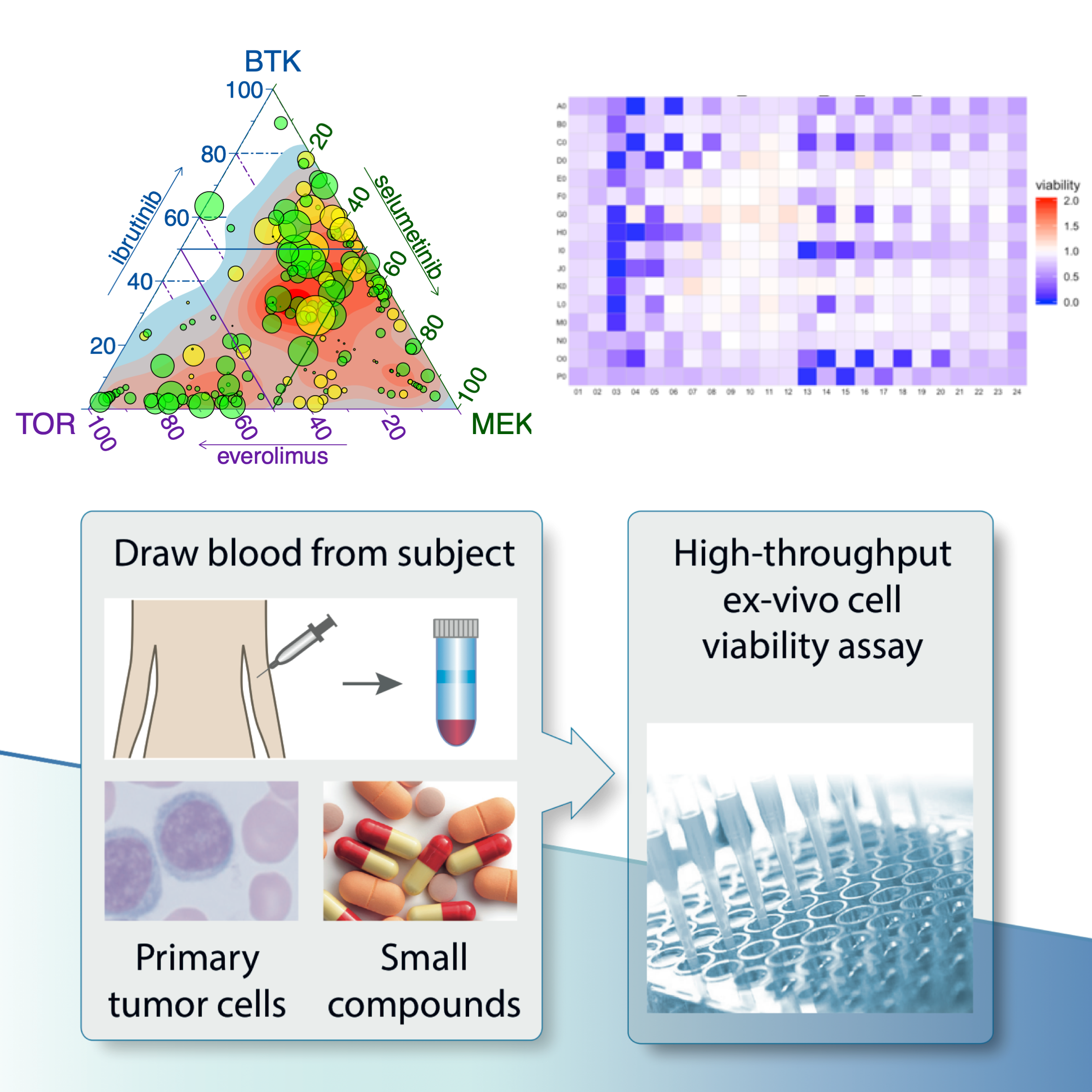
Functional Precision Medicine
Functional precision medicine is the approach that directly uses perturbation assays on primary patient samples to guide clinical decisions. We develop computational tools, in the forms of R/Bioconductor packages and Shiny apps, for streamlined processing, analyzing, and interpreting the results from functional assays, such as ex-vivo drug sensitivity assays, in order to increase their sensitivity and robustness, which are critical for clinical applications. Read a relevant study.
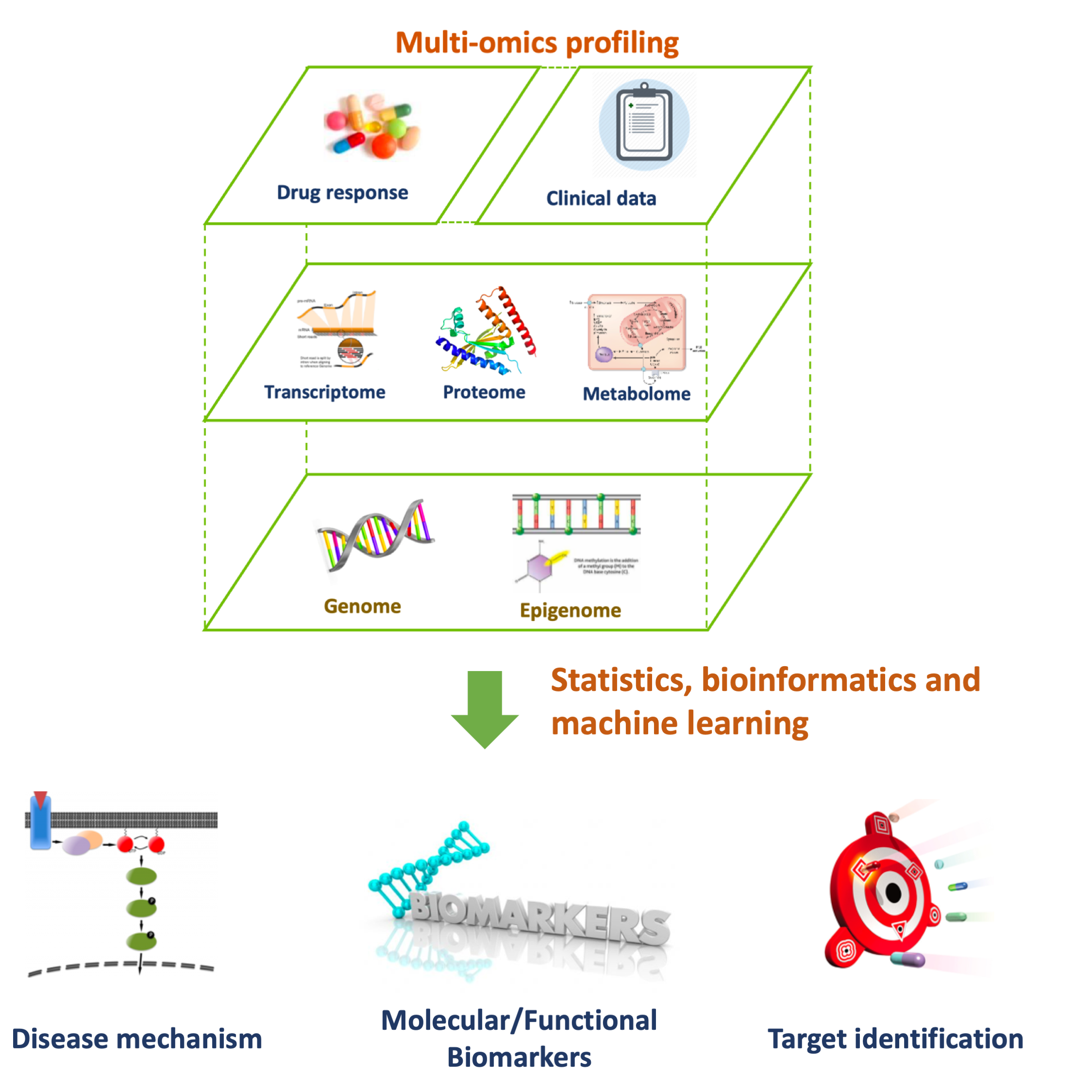
Multi-Omics & Machine Learning
Multi-omics integration is a powerful tool for understanding complex disease mechanisms. It also has unique advantages in clinical biomarker identification as it enables us to distinguish biological signals from incidental variation due to measurement noise or confounding experimental factors that tend to affect only individual data sources. We apply machine learning tools, such as multi-table factor analysis, for better integration and interpretation of multi-omics datasets. Read a relevant study.
Team Members

Junyan Lu, Ph.D
Research Group Leader
Dr. Junyan Lu studied Computational Biology and Drug Design at the Shanghai Institute of Materia Medica, Chinese Academy of Sciences. After obtaining his PhD in 2015, Dr. Lu joined Wolfgang Huber’s group at the European Molecular Biology Laboratory (EMBL) as a postdoc and later as a staff scientist, focusing on advancing personalized oncology of blood cancers through machine learning and multi-omics data integration. In December 2021, he joined the University Hospital Heidelberg and is currently leading a junior research group leader within the SMART-CARE consortium.

Giada Sandrini, PhD
Postdoctoral Researcher
Giada Sandrini earned her Master`s degree in Biomedical Engineering from Politecnico di Milano in 2019. She then completed her PhD in Computational Biomedicine at the Institute of Oncology Research in Bellinzona (CH), where she focused on employing multi-omics analysis to unravel molecular mechanisms and evolution in cancer. In July 2024 she joined the Lu group as Postdoctoral Researcher, to implement machine learning techniques on longitudinal proteomic and metabolomic data from lung cancer patients to improve tumor recurrence prediction.

Vladimir Vutov, PhD
Postdoctoral Researcher
Vladimir Vutov completed his BSc in Mathematical Statistics and his MSc in Econometrics at Sofia University, and his PhD in Mathematical Statistics at the University of Bremen. His doctoral thesis focuses on the development of novel analysis frameworks for addressing two relevant biological questions in the context of matrix-assisted laser desorption-ionization (MALDI) data. He joined the Lu group on May 1, 2024, as a Postdoc. His work is dedicated to the advancement of analysis methods tailored to mass spectrometry data.

Shubham Agrawal, M.Sc
Data Engineer / Technical Assistant
Shubham Agrawal obtained his Master’s degree in Computer Science from University of Freiburg in June 2022. He then joined Lu group on July 1st to work on the computational infrastructure for the MS data analysis pipeline in SMART-CARE.

Caroline Lohoff, M.Sc
PhD Candidate
Caroline Lohoff studied Molecular Biosciences with a focus on Systems and Computational Biology at Heidelberg University. After getting her Master’s degree in July 2022, she joined Lu group as a PhD student. Her PhD projects focus on identifying protein/metabolite-based biomarkers for tumor recurrence through mass spectrometry and multi-omics integration.

Qian-Wu Liao, M.Sc
PhD Candidate
Qian-Wu Liao obtained his Master’s degree in System Biology from Heidelberg University. He joined Lu group as a PhD student on September 1st, 2022, working on applying machine/deep learning models for identifying biomarkers for drug resistance and tumor recurrence.

Jiaojiao He, M.Sc
PhD Candidate
Jiajiao He got her Master’s degree in Bioinformatics and Systems Biology from the University of Amsterdam and Vrije Universiteit Amsterdam in August 2021. She joined Lu group as a PhD student on September 1st, 2022. Her PhD thesis focuses on developing and applying machine learning models to integrate omics and medical data for precision oncology.

Shuo Wang, M.Sc
Ph.D. Candidate
Shuo Wang is a Ph.D. candidate at Freiburg University. He joined Lu’s group on April 1st and is working on developing computational pipelines for processing and analyzing functional screen data.

Sasimonthakan Tanarsuwongkul, Pharm.D.
Ph.D. Candidate
Sasimonthakan received her Pharm.D. degree in Pharmaceutical Sciences from Chulalongkorn University, Thailand. She is pursuing a Ph.D. in Biochemistry at the University of South Carolina, USA. Her work focuses on deciphering signaling pathways using mass spectrometry-based proteomics and phosphoproteomics. She received a research grant from the German Academic Exchange Service (DAAD) and joined the Lu group in December 2023 to work on using multi-omics data integration to identify biomarkers of vascular calcification from plasma proteome and metabolome.
Alumni
Yueyang Xie was a master’s student at the Lu group between February 2022 and May 2023, working on normalization models for mass spectrometry-based phosphoproteomics data. She is currently a PhD student at University Hospital Heidelberg.
Luis Herfurth was an intern at the Lu group between December 2023 and February 2024, working on the development of pre-processing and visualization tools for targeted metabolomic data. He is currently a Bioinformatic research assistant at the Centre of Organismal Studies (COS), Heidelberg University.
Cong Quan Ta was a rotation student at the Lu Group between August and December 2023, working on implementing functional analysis modules for SMARTPhos, a tool for the analysis of phosphoproteomic data. He is currently a master’s student at DKFZ.
Miguel Hernandez was a rotation student at the Lu Group between February and April, working on the statistical analysis of ex-vivo and in-vivo drug responses from the SMARTrial study. He is currently a PhD student at the Institute for Computational Biology, Heidelberg University.

Get In Touch
- junyan.lu@uni-heidelberg.de
Drop By
Im Neuenheimer Feld 130.3, Room 10.318
69120 Heidelberg
Germany
